This is a reduced-median-median-joined calculation.
and the other paleolithic Ropers, which seem to form in two groups.
L. David Roper (roperld@vt.edu)
(www.roperld.com)
Back to Roper Y-Chromosome Project Main Page.
In a comparison of over 100 surname Y-chromosome markers I found that the closest surname to the Majority USA Roper family, also called the RoperY1 family, is the Campbell surname. Since then many Campbells have had their markers measured. Here I compare the Campbell markers with the paleolithic Roper markers.
The data are (Those ending in C and D are Campbells; those ending in R are Ropers:
| Allele (prefix DYS): |
393 | 390 | 19(394) | 391 | 385a | 385b | 426 | 388 | 439 | 389a | 392 | 389b | 458 | 459a | 459b | 455 | 454 | 447 | 437 | 448 | 449 | 464a | 464b | 464c | 464c | |
| 6304 | CoI | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 14 | 13 | 17 | 20 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 6361 | JAC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 20 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 7678 | AC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 20 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 31 | 15 | 15 | 16 | 17 |
| 6789 | PC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 20 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 31 | 15 | 15 | 15 | 17 |
| 9333 | JC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 4401 | WC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 6739 | HC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 29 | 15 | 15 | 16 | 17 |
| 6479 | JFC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 26 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 6341 | EC | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 13 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 428 | JDC | 12 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 7021 | JDC2 | 12 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 13 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 15 | 16 | 17 |
| 1566 | TBD | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 19 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 14 | 15 | 16 | 16 |
| 1245 | LDR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1247 | RSR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 2157 | NJR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 5647 | GTR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1248 | JHR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 16 |
| 1295 | CSR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 16 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1701 | SGR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 17 | 17 |
| 1250 | RPD | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 11 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 2155 | WAR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 18 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 5091 | GLR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 18 | 9 | 11 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1249 | DLR | 13 | 23 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 8449 | RJR | 13 | 23 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 5541 | RWR | 13 | 23 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 5940 | JER | 13 | 25 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 16 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 2156 | PSR | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 6084 | VKR | 13 | 24 | 14 | 10 | 11 | 15 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1294 | RLR | 12 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1299 | CFER | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 14 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1700 | DKR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 11 | 13 | 13 | 17 | 18 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 4868 | JWR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 14 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 5133 | RGR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 18 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 4867 | JJRj | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 13 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 8460 | DCR | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 26 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 8624 | JWR2 | 13 | 24 | 14 | 10 | 11 | 16 | 12 | 12 | 12 | 13 | 13 | 17 | 17 | 9 | 10 | 11 | 11 | 26 | 15 | 19 | 30 | 15 | 16 | 16 | 17 |
| 1296 | JJR | 13 | 23 | 14 | 10 | 11 | 14 | 12 | 12 | 12 | 13 | 13 | 17 | 18 | 9 | 10 | 11 | 11 | 24 | 15 | 19 | 30 | 15 | 15 | 17 | 17 |
| 1297 | LWR | 13 | 24 | 14 | 10 | 11 | 14 | 12 | 12 | 12 | 13 | 13 | 16 | 17 | 9 | 10 | 11 | 11 | 25 | 14 | 19 | 28 | 15 | 15 | 17 | 17 |
| 1606 | CMR | 13 | 24 | 14 | 11 | 11 | 14 | 12 | 12 | 11 | 13 | 13 | 16 | 18 | 8 | 10 | 11 | 11 | 25 | 15 | 19 | 29 | 14 | 15 | 17 | 17 |
| 1246 | JFHR | 13 | 23 | 14 | 11 | 11 | 15 | 12 | 12 | 12 | 13 | 14 | 16 | 17 | 9 | 10 | 11 | 11 | 24 | 15 | 19 | 30 | 15 | 15 | 17 | 17 |
| 2160 | GKR | 13 | 24 | 14 | 10 | 11 | 14 | 12 | 12 | 11 | 14 | 13 | 19 | 18 | 9 | 10 | 11 | 11 | 25 | 15 | 19 | 29 | 15 | 15 | 16 | 17 |
| 4146 | JRR | 13 | 24 | 14 | 11 | 11 | 13 | 12 | 13 | 12 | 14 | 13 | 16 | 17 | 9 | 9 | 11 | 11 | 25 | 15 | 19 | 29 | 15 | 15 | 15 | 17 |
DYS389I=DYS389a is subtracted from DYS389II to give DYS389b. If one does not do this one can get either one too many or one too few relative mutations. See Y-Chromosome Markers for Many Families.
The relative mutations among these individuals are:
| CoI | JAC | AC | PC | JC | WC | HC | JFC | EC | JDC | JDC2 | TBD | LDR | RSR | NJR | GTR | JHR | CSR | SGR | RPD | WAR | GLR | DLR | RJR | RWR | JER | PSR | VKR | RLR | CFER | DKR | JWR | RGR | JJRj | DCR | JWR2 | JJR | LWR | CMR | JFHR | GKR | JRR | ||
| CoI | 0 | CoI | |||||||||||||||||||||||||||||||||||||||||
| JAC | 1 | 0 | JAC | ||||||||||||||||||||||||||||||||||||||||
| AC | 2 | 1 | 0 | AC | |||||||||||||||||||||||||||||||||||||||
| PC | 3 | 2 | 1 | 0 | PC | ||||||||||||||||||||||||||||||||||||||
| JC | 2 | 1 | 2 | 3 | 0 | JC | |||||||||||||||||||||||||||||||||||||
| WC | 2 | 1 | 2 | 3 | 0 | 0 | WC | ||||||||||||||||||||||||||||||||||||
| HC | 3 | 2 | 2 | 3 | 1 | 1 | 0 | HC | |||||||||||||||||||||||||||||||||||
| JFC | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 0 | JFC | ||||||||||||||||||||||||||||||||||
| EC | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 2 | 0 | EC | |||||||||||||||||||||||||||||||||
| JDC | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 2 | 2 | 0 | JDC | ||||||||||||||||||||||||||||||||
| JDC2 | 4 | 3 | 4 | 5 | 2 | 2 | 3 | 3 | 1 | 1 | 0 | JDC2 | |||||||||||||||||||||||||||||||
| TBD | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 0 | TBD | ||||||||||||||||||||||||||||||
| LDR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 0 | LDR | |||||||||||||||||||||||||||||
| RSR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 0 | 0 | RSR | ||||||||||||||||||||||||||||
| NJR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 0 | 0 | 0 | NJR | |||||||||||||||||||||||||||
| GTR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 0 | 0 | 0 | 0 | GTR | ||||||||||||||||||||||||||
| JHR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | JHR | |||||||||||||||||||||||||
| CSR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 0 | CSR | ||||||||||||||||||||||||
| SGR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | SGR | |||||||||||||||||||||||
| RPD | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | RPD | ||||||||||||||||||||||
| WAR | 5 | 4 | 5 | 6 | 3 | 3 | 4 | 4 | 4 | 4 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 0 | WAR | |||||||||||||||||||||
| GLR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 1 | 1 | 0 | GLR | ||||||||||||||||||||
| DLR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 0 | DLR | |||||||||||||||||||
| RJR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | RJR | ||||||||||||||||||
| RWR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | 0 | RWR | |||||||||||||||||
| JER | 8 | 7 | 8 | 9 | 6 | 6 | 7 | 7 | 7 | 7 | 8 | 6 | 2 | 2 | 2 | 2 | 2 | 1 | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 0 | JER | ||||||||||||||||
| PSR | 5 | 4 | 5 | 6 | 3 | 3 | 4 | 4 | 4 | 4 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 0 | PSR | |||||||||||||||
| VKR | 5 | 4 | 5 | 6 | 3 | 3 | 4 | 4 | 4 | 4 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 0 | 0 | VKR | ||||||||||||||
| RLR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 4 | 5 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 0 | RLR | |||||||||||||
| CFER | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 0 | CFER | ||||||||||||
| DKR | 6 | 5 | 6 | 7 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 4 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 1 | 2 | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 3 | 0 | DKR | |||||||||||
| JWR | 5 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 0 | JWR | ||||||||||
| RGR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 2 | 0 | RGR | |||||||||
| JJRj | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 0 | JJRj | ||||||||
| DCR | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 4 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 0 | DCR | |||||||
| JWR2 | 7 | 6 | 7 | 8 | 5 | 5 | 6 | 4 | 6 | 6 | 7 | 5 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 0 | 0 | JWR2 | ||||||
| JJR | 7 | 6 | 7 | 7 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 6 | 7 | 7 | 7 | 7 | 7 | 8 | 6 | 8 | 6 | 7 | 6 | 6 | 6 | 9 | 6 | 6 | 8 | 8 | 7 | 8 | 8 | 8 | 8 | 8 | 0 | JJR | |||||
| LWR | 9 | 8 | 8 | 8 | 7 | 7 | 7 | 8 | 8 | 8 | 9 | 8 | 7 | 7 | 7 | 7 | 7 | 8 | 6 | 8 | 8 | 9 | 8 | 8 | 8 | 9 | 6 | 6 | 8 | 8 | 9 | 8 | 8 | 8 | 8 | 8 | 6 | 0 | LWR | ||||
| CMR | 11 | 10 | 10 | 10 | 9 | 9 | 8 | 10 | 10 | 10 | 11 | 8 | 10 | 10 | 10 | 10 | 10 | 11 | 9 | 11 | 9 | 10 | 11 | 11 | 11 | 12 | 9 | 9 | 11 | 11 | 8 | 11 | 11 | 11 | 11 | 11 | 8 | 7 | 0 | CMR | |||
| JFHR | 10 | 9 | 10 | 10 | 8 | 8 | 9 | 9 | 9 | 9 | 10 | 9 | 8 | 8 | 8 | 8 | 8 | 9 | 7 | 9 | 9 | 10 | 7 | 7 | 7 | 10 | 7 | 7 | 9 | 7 | 10 | 9 | 9 | 9 | 9 | 9 | 5 | 7 | 9 | 0 | JFHR | ||
| GKR | 7 | 8 | 8 | 9 | 7 | 7 | 6 | 8 | 8 | 8 | 9 | 8 | 9 | 9 | 9 | 9 | 9 | 10 | 9 | 10 | 8 | 9 | 10 | 10 | 10 | 11 | 8 | 8 | 10 | 10 | 7 | 8 | 8 | 10 | 10 | 10 | 8 | 9 | 8 | 13 | 0 | GKR | |
| JRR | 11 | 12 | 12 | 11 | 11 | 11 | 10 | 12 | 12 | 12 | 13 | 12 | 11 | 11 | 11 | 11 | 11 | 12 | 11 | 12 | 12 | 13 | 12 | 12 | 12 | 13 | 10 | 10 | 12 | 12 | 13 | 10 | 12 | 10 | 12 | 12 | 11 | 8 | 9 | 10 | 10 | 0 | JRR |
| CoI | JAC | AC | PC | JC | WC | HC | JFC | EC | JDC | JDC2 | TBD | LDR | RSR | NJR | GTR | JHR | CSR | SGR | RPD | WAR | GLR | DLR | RJR | RWR | JER | PSR | VKR | RLR | CFER | DKR | JWR | RGR | JJRj | DCR | JWR2 | JJR | LWR | CMR | JFHR | GKR | JRR |
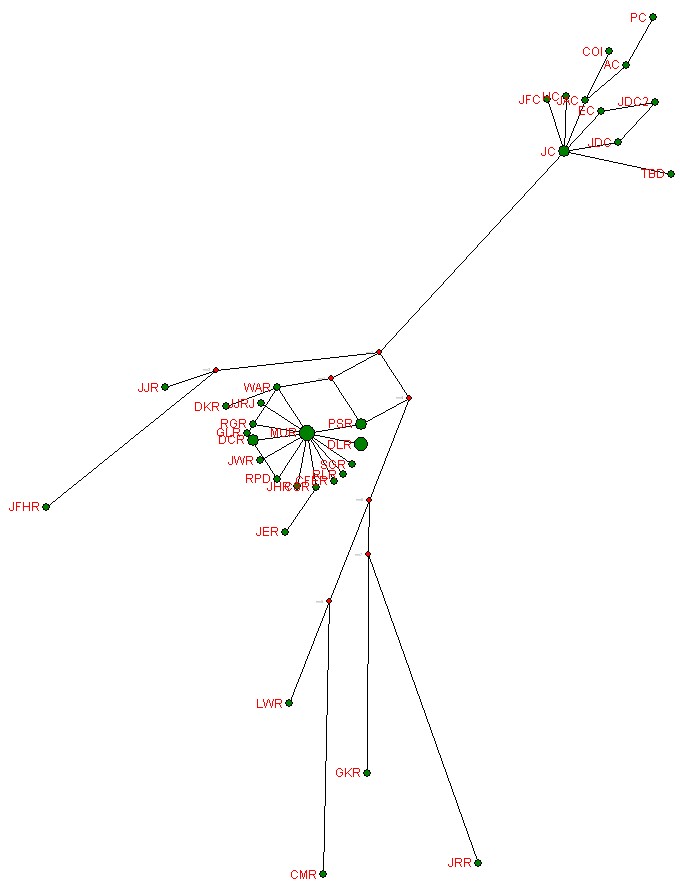
A way to look at the results is through a phylogenetic network. I use the free software of Fluxus Engineering (http://www.fluxus-engineering.com/sharenet.htm) to calculate and draw a network. (There are usually several possible networks for any given set of markers, but I present only the simplest looking one calculated by the program.) The phylogenetic network diagram is a visual way to see the genetic distances or relative mutations among the participants.
Another way to look at the results is through a phylogenetic network. I use the free software of Fluxus Engineering (http://www.fluxus-engineering.com/sharenet.htm) to calculate and draw a network. (The official brief instructions for using the program are at http://www.fluxus-engineering.com/netwinfaq.htm. I have some further instructions at Brief Instructions for Using the Phylogenetic Network Program)(There are usually several possible networks for any given set of markers, but I present only the simplest looking one calculated by the program.) The phylogenetic network diagram is a visual way to see the genetic distances or relative mutations among the participants.
A phylogenetic network for these individuals is:
 This is a reduced-median-median-joined calculation. |
| Note that the Campbells in the upper left corner are
quite distinct from the MUR/RoperY1 cluster and the other paleolithic Ropers, which seem to form in two groups. |
| This way of calculating a phylogenetic network does not account for the unusual nature of the four DYS464 markers. |
To create time-ordered phylograms of the various paleolithic groups, I use the PHYLIP Neighbor software, with the UPGMA option (Unweighted Pair Group Method with Arithmetic Mean), and the relative-mutations matrix to generate a tree file (*.tre) to be plotted by the TreeView software. (For a description of how I do it, see PHYLIPTreeViewUse.htm.)
Here is a time-ordered phylogram for the Ropers and the Campbells:
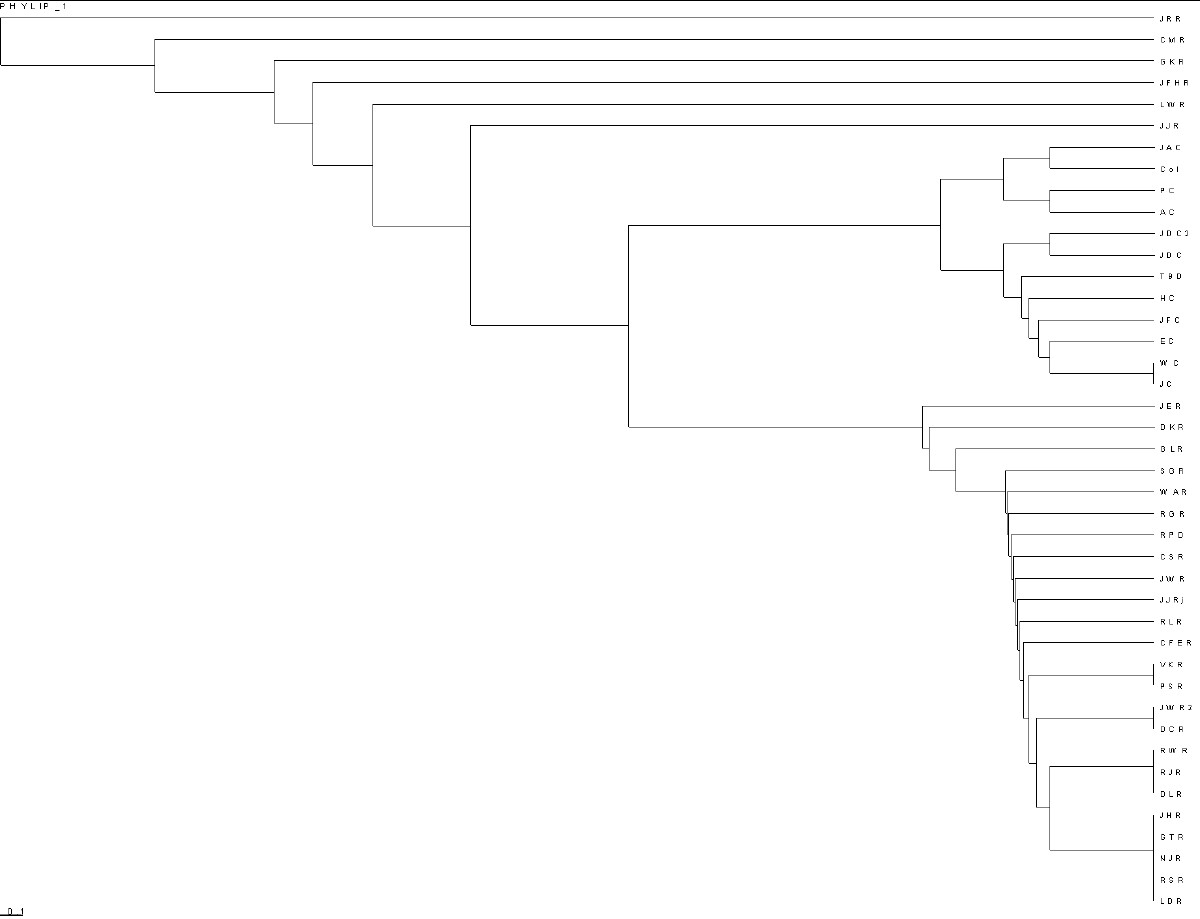 Present time is on the right. |
| Note that Campbells and the MUR/RoperY1s separated
from each other about 1200 ybp, after the other paleolithic Ropers separated from the common ancestors of all. This is about the time surnames came into use. |
| Here is another look at a phylogenetic network for the Campbells and paleolithic Ropers. |
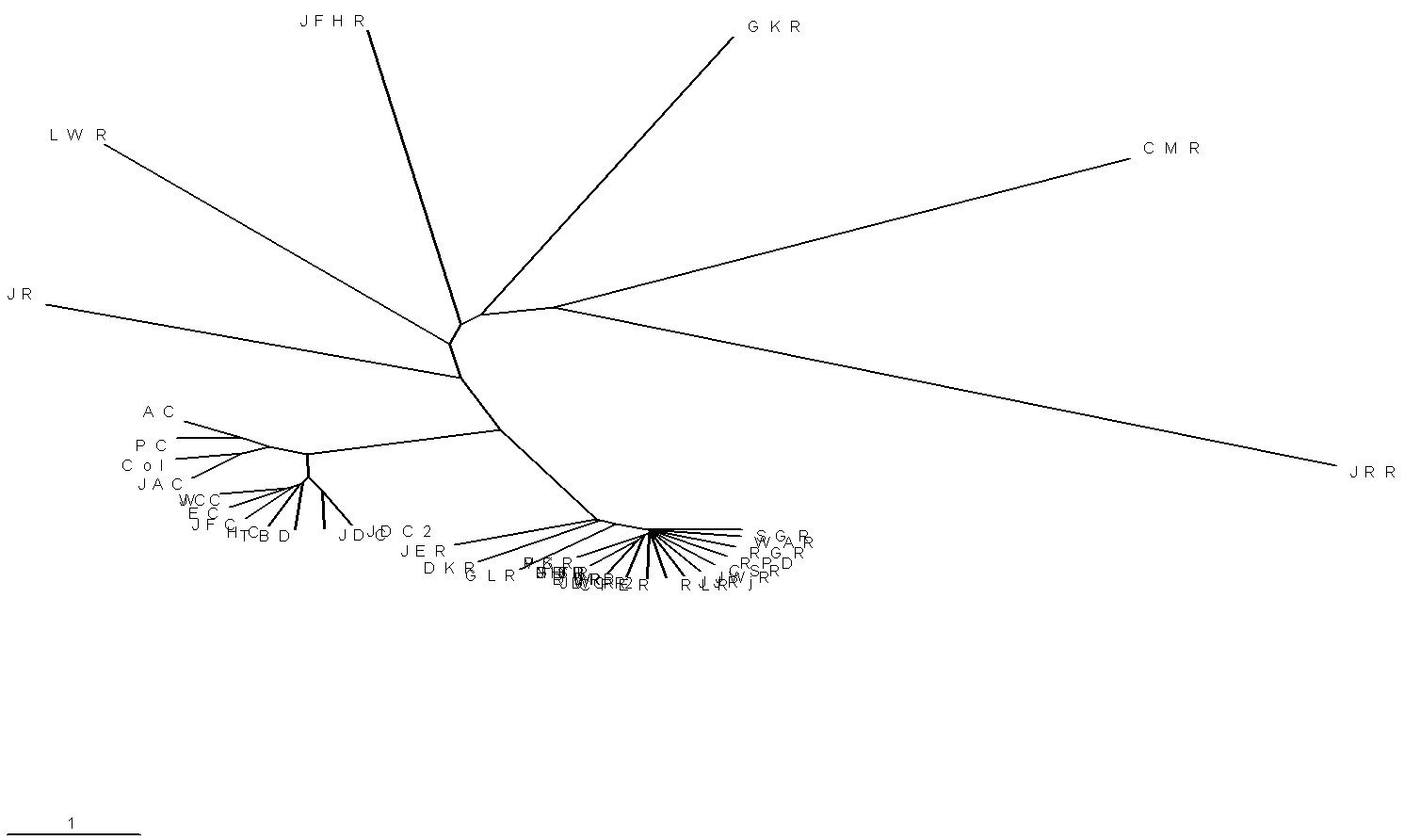 |
| This way of calculating a phylogenetic network does
account for the unusual nature of the four DYS464 markers, which shows better the close relationship between the Campbells and the MUR/RoperY1s. |